cKBET: assessing goodness of batch effect correction for single-cell RNA-seq
Por um escritor misterioso
Last updated 03 abril 2025

lt;p>Single-cell RNA sequencing reveals the gene structure and gene expression status of a single cell, which can reflect the heterogeneity between cells. However, batch effects caused by non-biological factors may hinder data integration and downstream analysis. Although the batch effect can be evaluated by visualizing the data, which actually is subjective and inaccurate. In this work, we propose a quantitative method cKBET, which considers the batch and cell type information simultaneously. The cKBET method accesses batch effects by comparing the global and local fraction of cells of different batches in different cell types. We verify the performance of our cKBET method on simulated and real biological data sets. The experimental results show that our cKBET method is superior to existing methods in most cases. In general, our cKBET method can detect batch effect with either balanced or unbalanced cell types, and thus evaluate batch correction methods.</p>
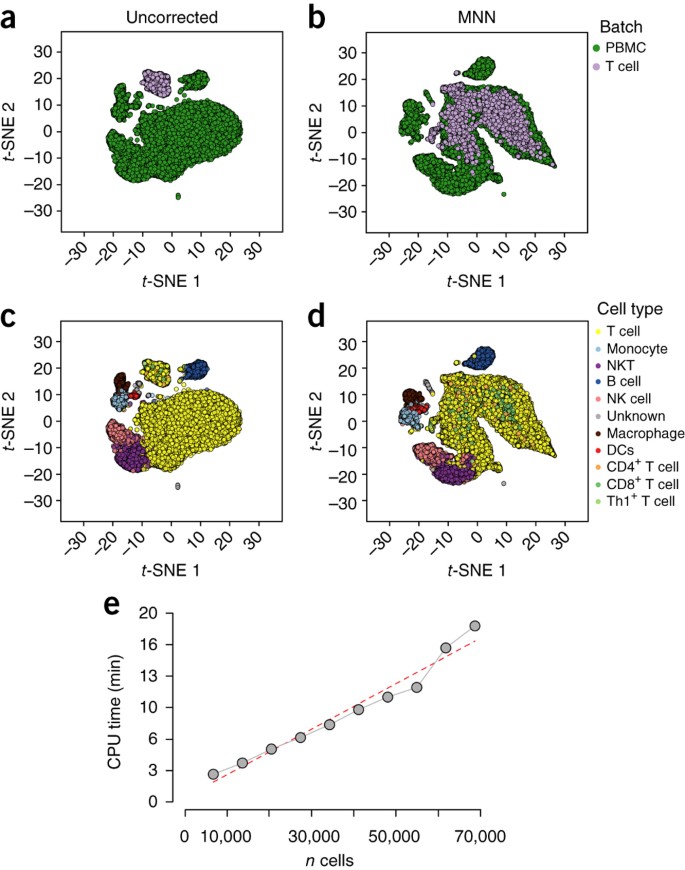
Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors

Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors
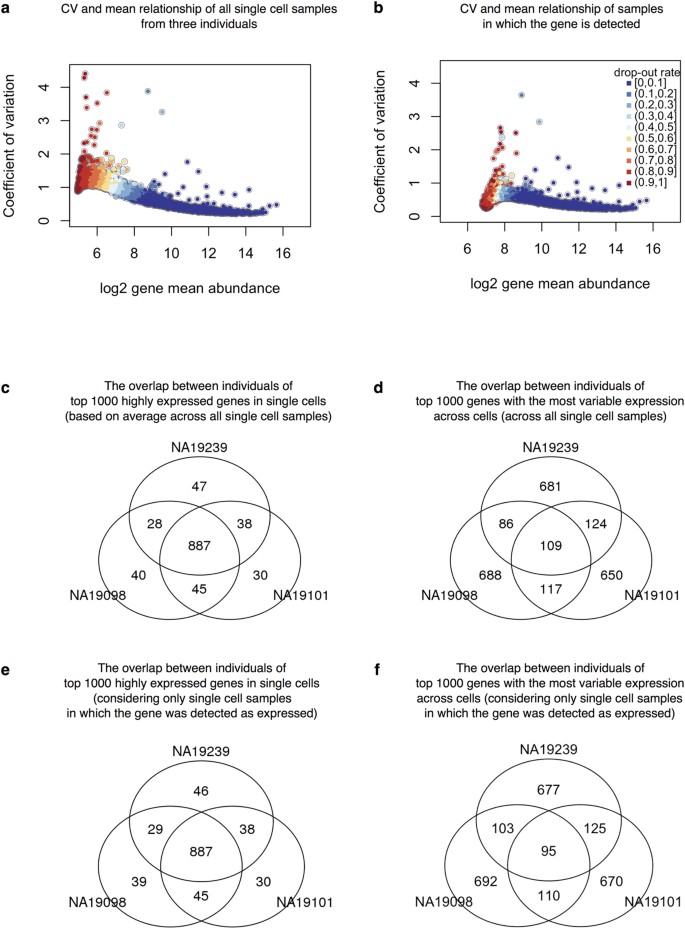
Batch effects and the effective design of single-cell gene expression studies

Assessment of batch-correction methods for scRNA-seq data with a new test metric

cKBET: assessing goodness of batch effect correction for single-cell RNA-seq

cKBET: assessing goodness of batch effect correction for single-cell RNA-seq
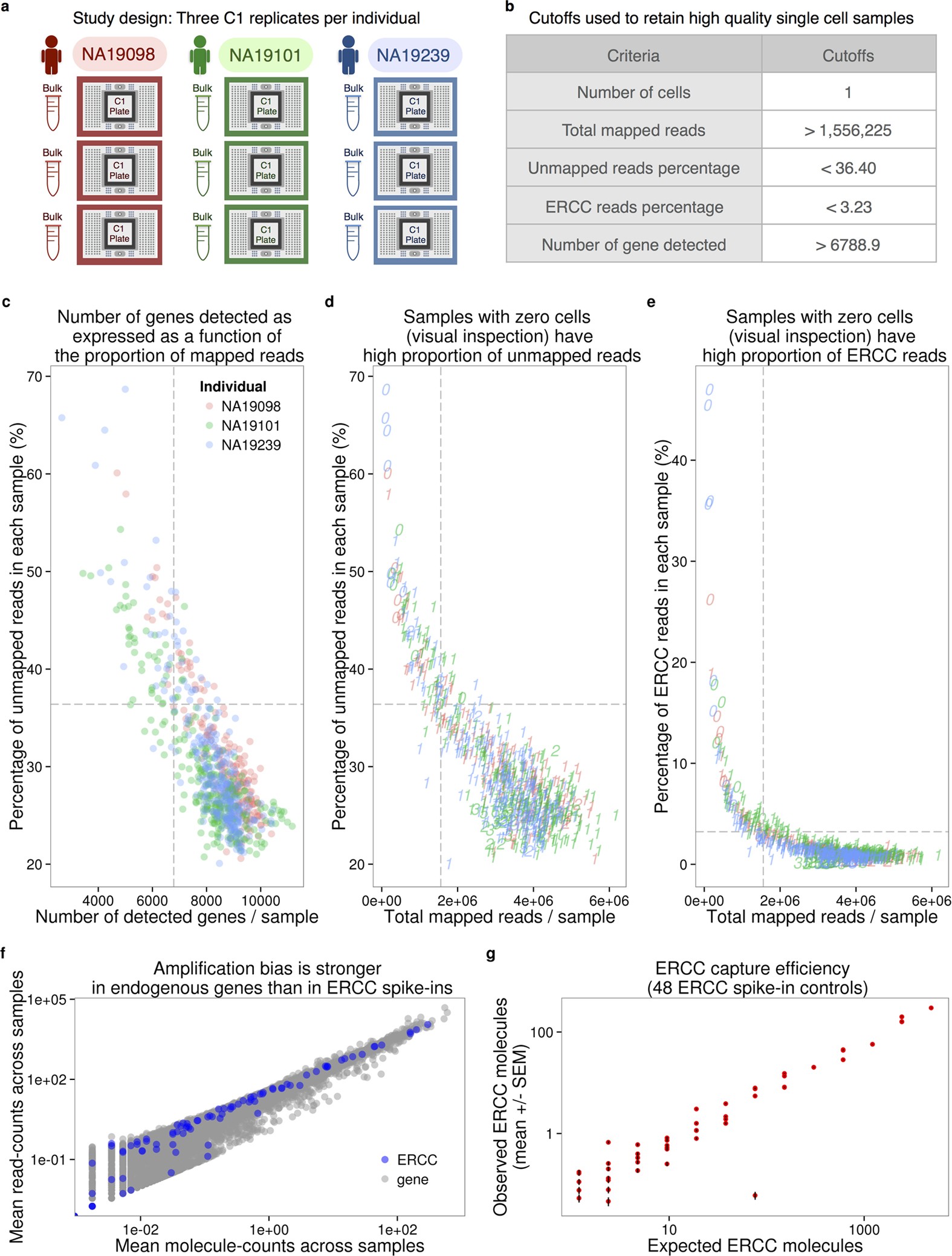
Batch effects and the effective design of single-cell gene expression studies
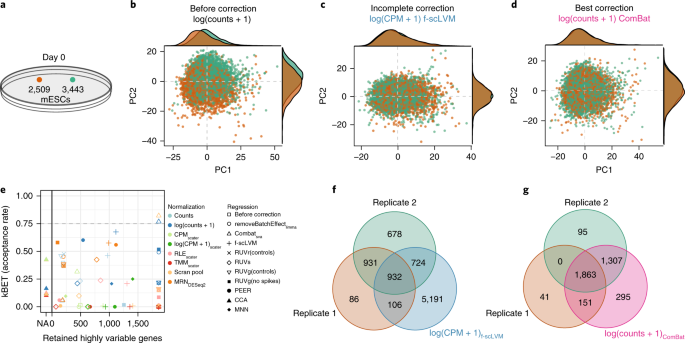
A test metric for assessing single-cell RNA-seq batch correction

Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors

PDF) A geometrical approach based on PCA to benchmark the algorithms of batch effect correction applied to the integration of RNA-Seq data
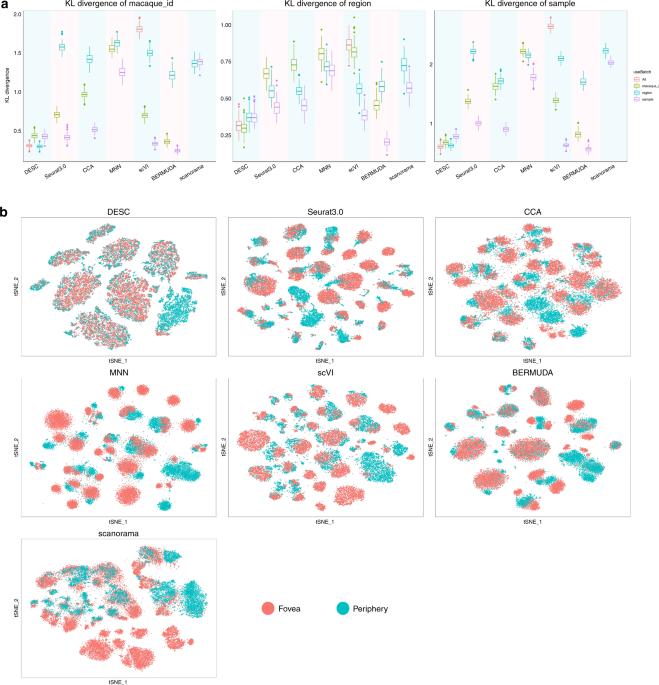
Deep learning enables accurate clustering with batch effect removal in single-cell RNA-seq analysis
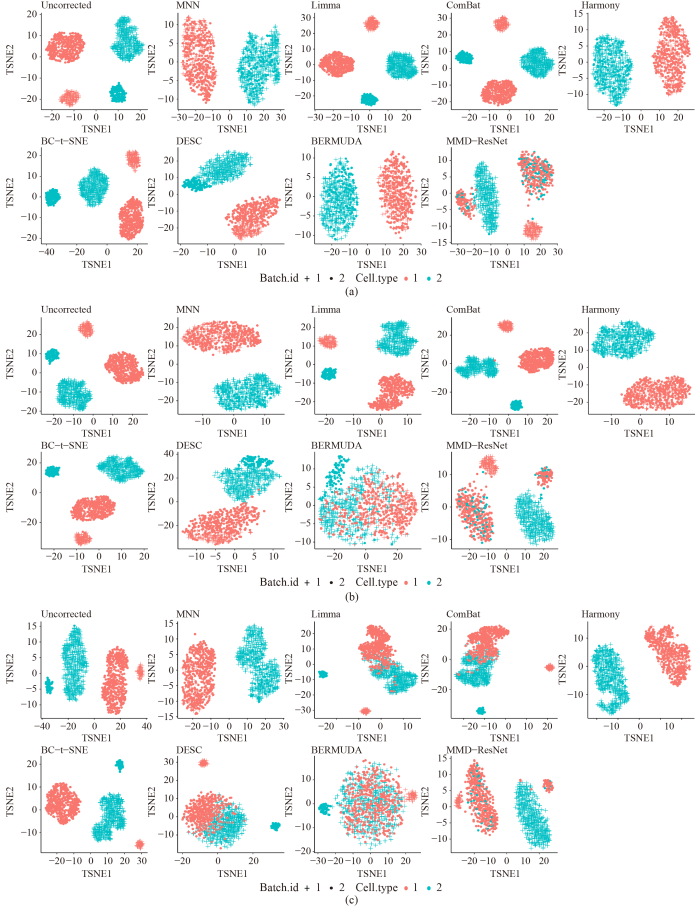
cKBET: assessing goodness of batch effect correction for single-cell RNA-seq

PDF) BEENE: Deep Learning based Nonlinear Embedding Improves Batch Effect Estimation

How to Batch Correct Single Cell. Comparing batch correction methods for…, by Nikolay Oskolkov
Recomendado para você
-
 A Plataforma de JOGOS DE CASSINO ONLINE Mais TOP e Mais Confiável!03 abril 2025
A Plataforma de JOGOS DE CASSINO ONLINE Mais TOP e Mais Confiável!03 abril 2025 -
 ckbet (FRAUDE)03 abril 2025
ckbet (FRAUDE)03 abril 2025 -
 ckbetcam (ckbet) - Replit03 abril 2025
ckbetcam (ckbet) - Replit03 abril 2025 -
 CKBet Cassino Online - Experiência emocionante em03 abril 2025
CKBet Cassino Online - Experiência emocionante em03 abril 2025 -
 ckbet – Fragment03 abril 2025
ckbet – Fragment03 abril 2025 -
 CKbet by imuninre198703 abril 2025
CKbet by imuninre198703 abril 2025 -
 CKBET APK for Android Download03 abril 2025
CKBET APK for Android Download03 abril 2025 -
 Ckbet - seu destino único para apostas online e jogos de cassino, Diário Arapiraca03 abril 2025
Ckbet - seu destino único para apostas online e jogos de cassino, Diário Arapiraca03 abril 2025 -
 HOME Ckbet03 abril 2025
HOME Ckbet03 abril 2025 -
 CKBET Paga Mesmo? Dá pra ganhar dinheiro?03 abril 2025
CKBET Paga Mesmo? Dá pra ganhar dinheiro?03 abril 2025
você pode gostar
-
JOIN THE DISCORD🔥-🔗 IN 🅱️ℹ️⭕ 1. The Ice Guy & His Cool Female Colle03 abril 2025
-
![Super Saiyan 4 Goku - Dragon Ball - Break Studio [IN STOCK]](https://www.relxelf.com/cdn/shop/files/218b60c6ded87cbfd584889da8c09ddf_800x.jpg?v=1690336364) Super Saiyan 4 Goku - Dragon Ball - Break Studio [IN STOCK]03 abril 2025
Super Saiyan 4 Goku - Dragon Ball - Break Studio [IN STOCK]03 abril 2025 -
 Tand Kids - Pote 40 Peças - Toyster Brinquedos - Toyster03 abril 2025
Tand Kids - Pote 40 Peças - Toyster Brinquedos - Toyster03 abril 2025 -
 Hell's Paradise in 14 minute - BiliBili03 abril 2025
Hell's Paradise in 14 minute - BiliBili03 abril 2025 -
 Scary-good deal: Microsoft Windows 10 only $13 in CdkeySales03 abril 2025
Scary-good deal: Microsoft Windows 10 only $13 in CdkeySales03 abril 2025 -
 Agora você pode jogar a demo de Like a Dragon: Ishin! no PC e Xbox03 abril 2025
Agora você pode jogar a demo de Like a Dragon: Ishin! no PC e Xbox03 abril 2025 -
 Cómo crear tu avatar en Roblox y personalizar tu personaje03 abril 2025
Cómo crear tu avatar en Roblox y personalizar tu personaje03 abril 2025 -
Como desenhar um animal fofo de desenho animado kawaii, Ecky O03 abril 2025
-
 Call of Duty: World At War — Braving The Frontlines Again As A 22-Year-Old, by Mirek Gosney, SUPERJUMP03 abril 2025
Call of Duty: World At War — Braving The Frontlines Again As A 22-Year-Old, by Mirek Gosney, SUPERJUMP03 abril 2025 -
 DanMachi Releases Season 3 Trailer!, Anime News03 abril 2025
DanMachi Releases Season 3 Trailer!, Anime News03 abril 2025

