Development of Moloney Murine Leukemia Virus Reverse Transcriptase Fused with Archaeal DNA-binding Protein Sis7a
Por um escritor misterioso
Last updated 28 março 2025


Comparison of the salt tolerance of Sso7d fusions and the unmodified

Characterization of a fusion NeqSSB-TaqS DNA polymerase in comparison

Development of Moloney Murine Leukemia Virus Reverse Transcriptase Fused with Archaeal DNA-binding Protein Sis7a

Graphical presentation of the relative adaptiveness (wij) of the codon

Activity of the M-MuLV RT point mutants. The 50 8 /37 8 C activity

Distribution of CAI values of the C.elegans genome under different

HADDOCK web server statistics (as of September 1st 2015). The worldwide
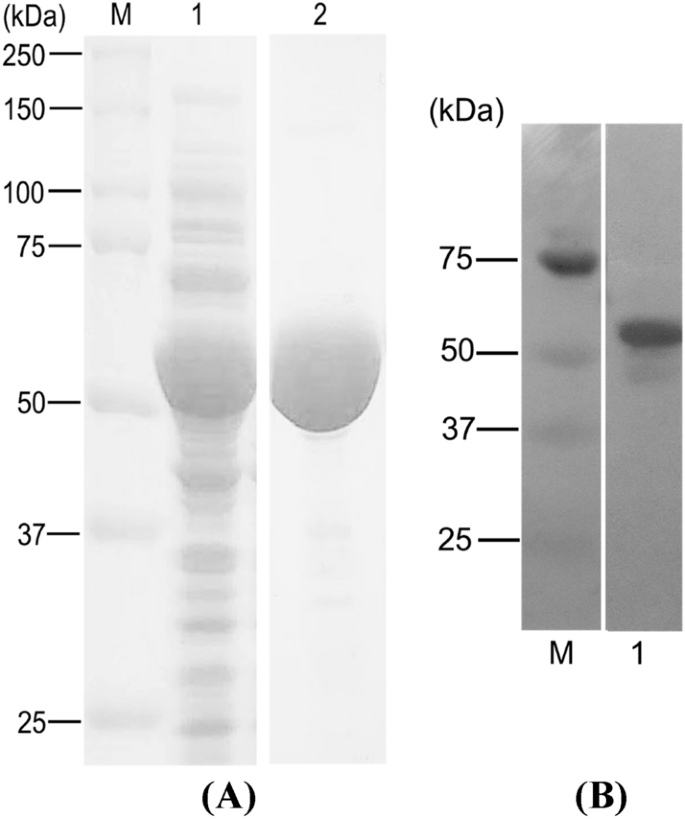
Expression of Codon-Optimized Gene Encoding Murine Moloney Leukemia Virus Reverse Transcriptase in Escherichia coli

The NiceProt view of a SWISS-PROT entry presents its contents in a
Recomendado para você
-
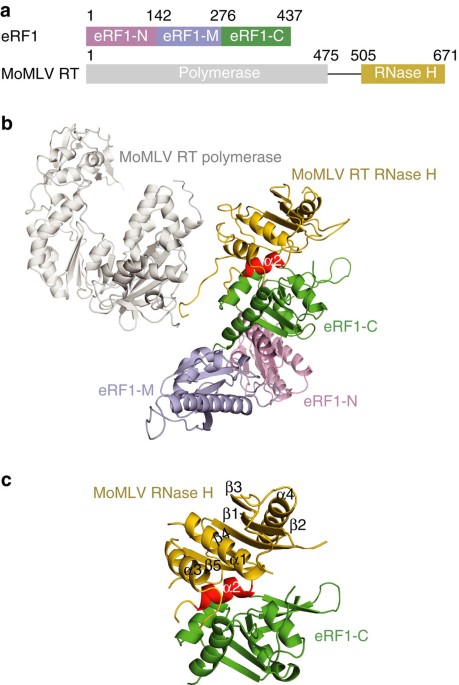 Structural basis of suppression of host translation termination by Moloney Murine Leukemia Virus28 março 2025
Structural basis of suppression of host translation termination by Moloney Murine Leukemia Virus28 março 2025 -
 Gg打xg的两二把,说明这版本的团队装真的必须要有! 17828 março 2025
Gg打xg的两二把,说明这版本的团队装真的必须要有! 17828 março 2025 -
 SNID11 quebra recordes na estreia e tem maior liquidez do setor28 março 2025
SNID11 quebra recordes na estreia e tem maior liquidez do setor28 março 2025 -
 Reverse Transcriptase of Moloney Murine Leukemia Virus Binds to Eukaryotic Release Factor 1 to Modulate Suppression of Translational Termination: Cell28 março 2025
Reverse Transcriptase of Moloney Murine Leukemia Virus Binds to Eukaryotic Release Factor 1 to Modulate Suppression of Translational Termination: Cell28 março 2025 -
 SNID11 quebra recordes na listagem na B3 e tem o maior giro diário do setor28 março 2025
SNID11 quebra recordes na listagem na B3 e tem o maior giro diário do setor28 março 2025 -
 Avian Myeoblastosis Virus (AMV) Moloney Murine Leukemia Virus (MMLV) , HIV Reverse Transcriptase28 março 2025
Avian Myeoblastosis Virus (AMV) Moloney Murine Leukemia Virus (MMLV) , HIV Reverse Transcriptase28 março 2025 -
 Safety Recall RVXX1904 - I-Shift Transmission Auxiliary Air Tank - 2019-2020 Volvo VAH, VHD, VNL, & VNR28 março 2025
Safety Recall RVXX1904 - I-Shift Transmission Auxiliary Air Tank - 2019-2020 Volvo VAH, VHD, VNL, & VNR28 março 2025 -
 Muv-Luv Alternative The Animation Sound Track, Muv-Luv Wiki28 março 2025
Muv-Luv Alternative The Animation Sound Track, Muv-Luv Wiki28 março 2025 -
 MV Sport Corded Crewneck Pullover28 março 2025
MV Sport Corded Crewneck Pullover28 março 2025 -
 m1v – blood Lyrics28 março 2025
m1v – blood Lyrics28 março 2025
você pode gostar
-
 Linha arte olho desenho cabelo, olho, branco, cara, pessoas png28 março 2025
Linha arte olho desenho cabelo, olho, branco, cara, pessoas png28 março 2025 -
Anime News And Facts on X: Konosuba Upcoming new anime will reveal new information on May 28, 2022. / X28 março 2025
-
 Fastest way to farm Stone! (Episode Adventure: Majin Buu Saga (Z))28 março 2025
Fastest way to farm Stone! (Episode Adventure: Majin Buu Saga (Z))28 março 2025 -
 The God of High School, Korean Webtoons Wiki28 março 2025
The God of High School, Korean Webtoons Wiki28 março 2025 -
 Afterlife Lyrics28 março 2025
Afterlife Lyrics28 março 2025 -
 AUTOCLICKER VS PRO LOBBIES in roblox blade ball28 março 2025
AUTOCLICKER VS PRO LOBBIES in roblox blade ball28 março 2025 -
 Julie Van Der Wielen – Instituto de Filosofía UDP28 março 2025
Julie Van Der Wielen – Instituto de Filosofía UDP28 março 2025 -
 Sonic vs Shadow coloring pages/ Sonic, Silver, Amy Rose coloring28 março 2025
Sonic vs Shadow coloring pages/ Sonic, Silver, Amy Rose coloring28 março 2025 -
 Outer Banks Season 2 Netflix Release Date, Cast And Plot - What We28 março 2025
Outer Banks Season 2 Netflix Release Date, Cast And Plot - What We28 março 2025 -
 Na abertura das olimpíadas, Mananciais lança música do Colégio28 março 2025
Na abertura das olimpíadas, Mananciais lança música do Colégio28 março 2025
